6.3 Examples
The following example show how these concepts can be applied in practice.
6.3.1 tidy up flatTempHum.csv
6.3.1.1 Loading data and parsing timestamp
library(tidyverse)
library(lubridate)
# load test data set
df <- read.csv("https://github.com/hslu-ige-laes/edar/raw/master/sampleData/flatTempHum.csv",
stringsAsFactors=FALSE,
sep =";")
df$time <- parse_date_time(df$time, "YmdHMS", tz = "Europe/Zurich")
head(df, 5)## time FlatA_Hum FlatA_Temp FlatB_Hum FlatB_Temp FlatC_Hum
## 1 2018-10-03 00:00:00 53.0 24.43 38.8 22.40 44.0
## 2 2018-10-03 01:00:00 53.0 24.40 38.8 22.40 44.0
## 3 2018-10-03 02:00:00 53.0 24.40 39.3 22.40 44.7
## 4 2018-10-03 03:00:00 53.0 24.40 40.3 22.40 45.0
## 5 2018-10-03 04:00:00 53.3 24.40 41.0 22.37 45.2
## FlatC_Temp FlatD_Hum FlatD_Temp
## 1 24.5 49.0 24.43
## 2 24.5 49.0 24.40
## 3 24.5 48.3 24.38
## 4 24.5 48.0 24.33
## 5 24.5 47.7 24.306.3.1.2 tidy up
What needs to be done here to get this data set into a tidy format?
- Firstly each observation should have its own row… To fulfill that, we have to convert from wide to long.
- Secondly, the variables are
flatandsensorwhich are currently merged together in the column name. That must be separated.
# convert wide to long format
df.long <- as.data.frame(tidyr::pivot_longer(df,
cols = -time,
names_to = "datapoint",
values_to = "value",
values_drop_na = TRUE))
head(df.long, 5)## time datapoint value
## 1 2018-10-03 FlatA_Hum 53.00
## 2 2018-10-03 FlatA_Temp 24.43
## 3 2018-10-03 FlatB_Hum 38.80
## 4 2018-10-03 FlatB_Temp 22.40
## 5 2018-10-03 FlatC_Hum 44.00# separate datapoint into two columns
df.separated <- df.long %>%
separate(col = datapoint, into = c("flat", "sensor"), sep = "_") %>%
mutate_at("flat", str_replace, "Flat", "") %>%
na.omit()
head(df.separated, 10)## time flat sensor value
## 1 2018-10-03 00:00:00 A Hum 53.00
## 2 2018-10-03 00:00:00 A Temp 24.43
## 3 2018-10-03 00:00:00 B Hum 38.80
## 4 2018-10-03 00:00:00 B Temp 22.40
## 5 2018-10-03 00:00:00 C Hum 44.00
## 6 2018-10-03 00:00:00 C Temp 24.50
## 7 2018-10-03 00:00:00 D Hum 49.00
## 8 2018-10-03 00:00:00 D Temp 24.43
## 9 2018-10-03 01:00:00 A Hum 53.00
## 10 2018-10-03 01:00:00 A Temp 24.40This dataset can now be considered tidy and is ready for further processing.
6.3.1.3 convert to ‘tsibble’
library(tsibble)
# convert the data frame in a tsibble object
tsbl <- as_tsibble(df.separated, key = c(flat, sensor), index = time)
tsbl## # A tsibble: 131,602 x 4 [1h] <Europe/Zurich>
## # Key: flat, sensor [8]
## time flat sensor value
## <dttm> <chr> <chr> <dbl>
## 1 2018-10-03 00:00:00 A Hum 53
## 2 2018-10-03 01:00:00 A Hum 53
## 3 2018-10-03 02:00:00 A Hum 53
## 4 2018-10-03 03:00:00 A Hum 53
## 5 2018-10-03 04:00:00 A Hum 53.3
## 6 2018-10-03 05:00:00 A Hum 53.7
## 7 2018-10-03 06:00:00 A Hum 50.8
## 8 2018-10-03 08:00:00 A Hum 42.7
## 9 2018-10-03 09:00:00 A Hum 46
## 10 2018-10-03 10:00:00 A Hum 41.3
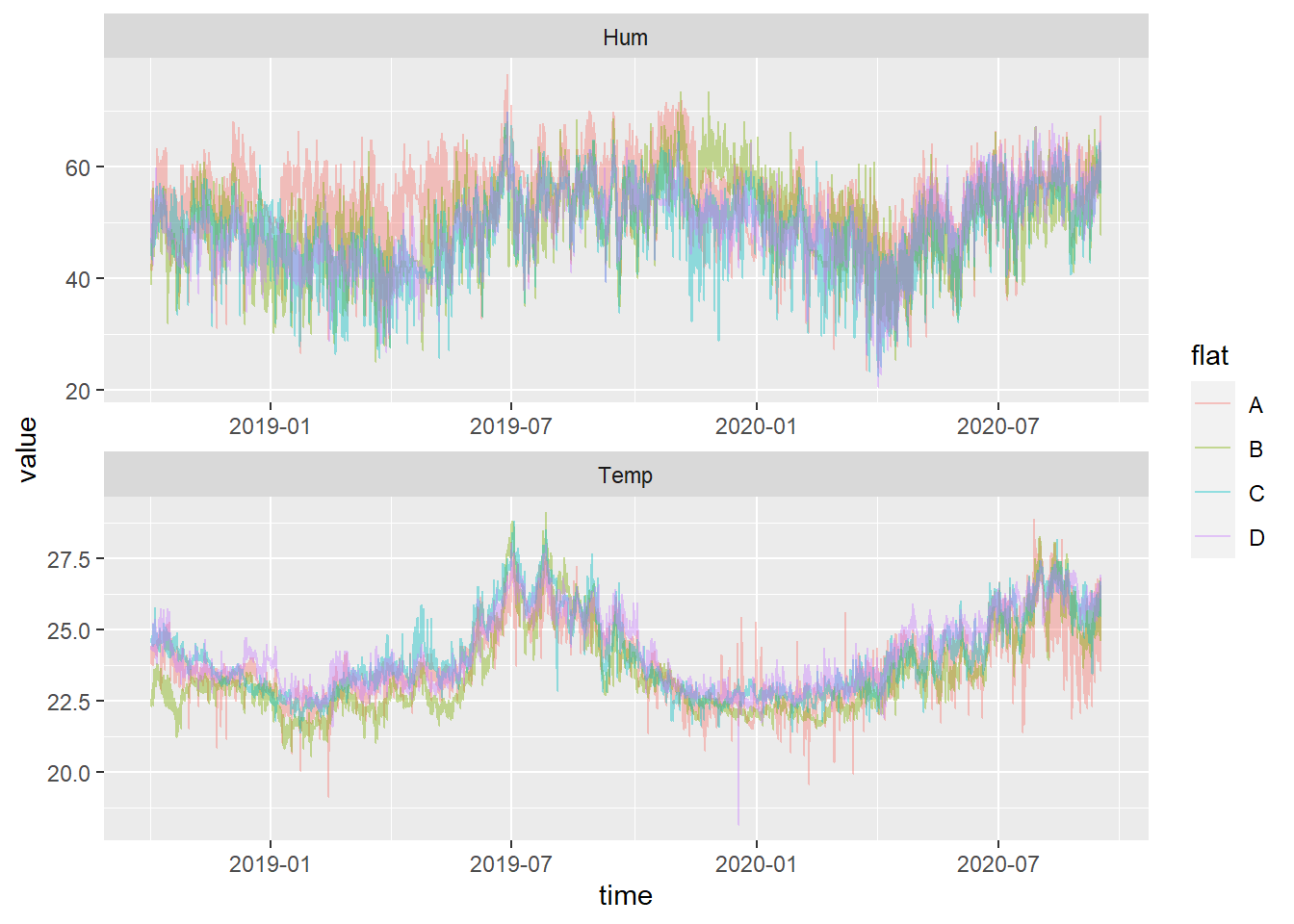
## # ... with 131,592 more rows6.3.1.4 Visualization
It’s now relatively easy and straightforward to make a plot using the key-columns flat and sensor:
library(ggplot2)
ggplot(tsbl, aes(x= time, y= value, colour = flat)) +
geom_line(alpha=0.4) +
facet_wrap(~ sensor, scales="free", ncol = 1)